Integrated 3D Tomography and Computational Modeling to Study Forces in Metaphase Spindles
Abstract
Chromosome segregation during mitosis and meiosis are fundamental processes of life. During its course a bipolar spindle assembles and initially aligns the chromosomes on a metaphase plate before they are segregated to the two arising daughter cells during anaphase. The different cellular components of mitotic and meiotic spindles: microtubules, chromosomes, motor proteins and also membranes, such as the cell cortex or nuclear envelope, must interact with each other during spindle assembly, chromosome alignment and segregation.
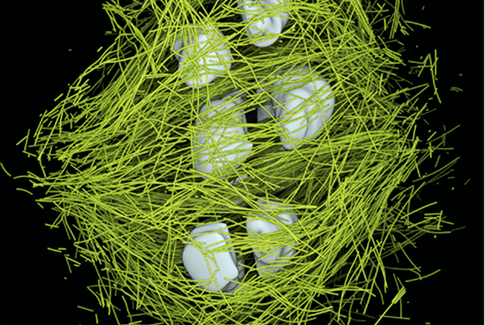
While we have achieved a very good understanding of signaling cascades and checkpoints that monitor and control mitosis, we are lacking an understanding on how the nano‐scale components of a spindle physically interact and generate the forces needed to faithfully align and segregate chromosomes. We have very little knowledge about the mechanics of spindles. In order to overcome the current limitations, we are generating a holistic description of mitotic and meiotic spindles in C. elegans embryos and mouse oocytes. This ambitious task will be achieved by a combination of 3D electron tomography, light microscopy and computational simulations. 3D tomography provides unprecedented detailed information about every individual microtubule within the spindle. We can measure the length, position and interactions of single microtubules with subcellular targets. In addition, we have developed tools to analyze the curvature of microtubules. Curvature originates from forces experienced by the microtubule and provides a read out of spindle intrinsic forces. The source of those forces can be determined by a combined approach of mutant analysis and computational simulations. To complement the static tomography data, we perform state‐of‐the‐art light microscopy. In conjunction with the light and electron microscopy we develop hypothesis and mechanical models of chromosome congression, alignment and segregation and test those in computational simulations and by selective depletion of proteins. We are using this approach to develop a (near) true‐to‐life model of microtubule, chromosome and membrane interactions and forces during mitosis.